Abstract
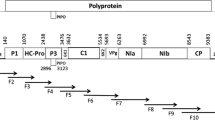
Disease outbreaks caused by turnip yellows virus (TuYV), a member of the genus Polerovirus, family Luteoviridae, regularly occur in canola and pulse crops throughout Australia. To understand the genetic diversity of TuYV for resistance breeding and management, genome sequences of 28 TuYV isolates from different hosts and locations were determined using high-throughput sequencing (HTS). We aimed to identify the parts of the genome that were most variable and clarify the taxonomy of viruses related to TuYV. Poleroviruses contain seven open reading frames (ORFs): ORF 0–2, 3a, and 3–5. Phylogenetic analysis based on the genome sequences, including isolates of TuYV and brassica yellows virus (BrYV) from the GenBank database, showed that most genetic variation among isolates occurred in ORF 5, followed by ORF 0 and ORF 3a. Phylogenetic analysis of ORF 5 revealed three TuYV groups; P5 group 1 and group 3 shared 45–49% amino acid sequence identity, and group 2 is a recombinant between the other two. Phylogenomic analysis of the concatenated ORFs showed that TuYV is paraphyletic with respect to BrYV, and together these taxa form a well-supported monophyletic group. Our results support the hypothesis that TuYV and BrYV belong to the same species and that the phylogenetic topologies of ORF 0, 3a and 5 are incongruent and may not be informative for species demarcation. A number of beet western yellow virus (BWYV)- and TuYV-associated RNAs (aRNA) were also identified by HTS for the first time in Australia.





Similar content being viewed by others
References
Abraham AD, Varrelmann M, Vetten HJ (2008) Molecular evidence for the occurrence of two new luteoviruses in cool season food legumes in Northeast Africa. Afr J Biotechnol 7:414–420
Asaad NY, Kumari SG, Haj-Kassem AA, Shalaby A-BA, Al-Shaabi S, Malhotra RS (2009) Detection and characterization of Chickpea Chlorotic Stunt Virus in Syria. J Phytopathol 157:756–761
Asare-Bendiako E (2011) Brassicaceae: turnip yellows virus interactions. Doctoral dissertation, University of Warwick, Coventry. http://webcat.warwick.ac.uk/record=b2553527~S1
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Baumberger N, Tsai C-H, Lie M, Havecker E, Baulcombe David C (2007) The polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17:1609–1614
Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V (2007) The polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol 17:1615–1621
Brault V, van den Heuvel JF, Verbeek M, Ziegler-Graff V, Reutenauer A, Herrbach E, Garaud JC, Guilley H, Richards K, Jonard G (1995) Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J 14:650–659
Brault V, Mutterer J, Scheidecker D, Simonis MT, Herrbach E, Richards K, Ziegler-Graff V (2000) Effects of point mutations in the readthrough domain of the beet western yellows virus minor capsid protein on virus accumulation in planta and on transmission by aphids. J Virol 74:1140–1148
Brault V, Périgon S, Reinbold C, Erdinger M, Scheidecker D, Herrbach E, Richards K, Ziegler-Graff V (2005) The polerovirus minor capsid protein determines vector specificity and intestinal tropism in the aphid. J Virol 79:9685–9693
Bushnell B (2016) BBTools: a suit of bioinformatic tools used for DNA and RNA sequence data analysis. [WWW document] URL http://jgi.doe.gov/data-and-tools/bbtools/.
Chin L-S, Foster JL, Falk BW (1993) The beet western yellows virus ST9-associated RNA shares structural and nucleotide sequence homology with carmo-like viruses. Virology 192:473–482
Congdon B, Matson P, Begum F, Kehoe M, Coutts B (2019) Application of loop-mediated isothermal amplification in an early warning system for epidemics of an externally sourced plant virus. Plants 8:139
Congdon BS, Kehoe MA, Filardo FF, Coutts BA (2019) In-field capable loop-mediated isothermal amplification detection of Turnip yellows virus in plants and its principal aphid vector Myzus persicae. J Virol Methods 265:15–21
Coutts BA, Hawkes JR, Jones RAC (2006) Occurrence of Beet western yellows virus and its aphid vectors in over-summering broad-leafed weeds and volunteer crop plants in the grainbelt region of south-western Australia. Aust J Agric Res 57:975–982
Coutts BA, Jones RAC, Umina P, Davidson J, Baker G, Aftab M (2015) Beet western yellows virus (Synon: Turnip yellows virus) and green peach aphid in canola. South Australian GRDC Updates, Canberra, Australia. https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2015/02/beet-western-yellows-virus-synonym-turnip-yellows-virus-and-green-peach-aphid-in-canola
DeBlasio SL, Johnson R, Mahoney J, Karasev A, Gray SM, MacCoss MJ, Cilia M (2015) insights into the polerovirus-plant interactome revealed by coimmunoprecipitation and mass spectrometry. Mol Plant-Microbe Interact 28:467–481
Dombrovsky A, Glanz E, Lachman O, Sela N, Doron-Faigenboim A, Antignus Y (2013) The complete genomic sequence of pepper yellow leaf curl virus (PYLCV) and its implications for our understanding of evolution dynamics in the genus polerovirus. PLoS ONE 8:e70722–e70722
Domingo E, Baranowski E, Ruiz-Jarabo CM, Martín-Hernández AM, Sáiz JC, Escarmís C (1998) Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis 4:521–527
Duffus JE, Russell GE (1970) Serological and host range evidence for the occurence of beet western yellows virus in Europe. Phytopathology 60:1199–1202
Falk BW, Chin L-S, Duffus JE (1989) Complementary DNA cloning and hybridization analysis of beet western yellows luteovirus RNAs. J Gen Virol 70:1301–1309
Falk BW, Duffus JE (1984) Identification of small single- and double-stranded RNAs associated with severe symptoms in beet western yellows virus-infected capsella bursa-pastoris. Phytopathology 74:1224–1229
Fiallo-Olivé E, Navas-Hermosilla E, Ferro CG, Zerbini FM, Navas-Castillo J (2018) Evidence for a complex of emergent poleroviruses affecting pepper worldwide. Arch Virol 163:1171–1178
Filardo FF, Sharman M (2019) Siratro latent polerovirus (SLPV): a new polerovirus from Australia with a non-functional open reading frame 0. Australas Plant Path 48:491–501
Filardo FF, Thomas JE, Webb M, Sharman M (2019) Faba bean polerovirus 1 (FBPV-1); a new polerovirus infecting legume crops in Australia. Arch Virol 164:1915–1921
Freeman A, Aftab M (2011) Effective management of viruses in pulse crops in south eastern Australia should include management of weeds. Australas Plant Path 40:430–441
Gray S, Cilia M, Ghanim M (2014) Chapter four - circulative, “nonpropagative” virus transmission: an orchestra of virus-, insect-, and plant-derived instruments. In: Maramorosch K, Murphy FA (eds) Advances in virus research. Academic Press, Cambridge, pp 141–199
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Harrison BD (2002) Virus variation in relation to resistance-breaking in plants. Euphytica 124:181–192
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2017) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522
Huson DH, Bryant D (2005) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Jaag HM, Kawchuk L, Rohde W, Fischer R, Emans N, Prüfer D (2003) An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. P Natl Acad Sci 100:8939–8944
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8:275–282
Katul L (1992) Characterization by serology and molecular biology of bean leaf roll virus and faba bean necrotic yellows virus. PhD thesis, University of Göttingen, Göttingen, Germany, p 115
King AM, Lefkowitz E, Adams MJ, Carstens EB (2011) Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier, Amsterdam
Kinoti WM, Nancarrow N, Dann A, Rodoni BC, Constable FE (2020) Updating the quarantine status of prunus infecting viruses in Australia. Viruses 12:246
Kruger F (2015) Trim Galore. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
Kuhn JH, Wolf YI, Krupovic M, Zhang Y-Z, Maes P, Dolja VV, Koonin EV (2019) Classify viruses—the gain is worth the pain. Nature 566:318–320
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Lim S, Yoo RH, Igori D, Zhao F, Kim KH, Moon JS (2015) Genome sequence of a recombinant brassica yellows virus infecting Chinese cabbage. Arch Virol 160:597–600
Liu M, Liu X, Li X, Zhang D, Dai L, Tang Q (2016) Complete genome sequence of a Chinese isolate of pepper vein yellows virus and evolutionary analysis based on the CP, MP and RdRp coding regions. Arch Virol 161:677–683
Lotos L, Olmos A, Orfanidou C, Efthimiou K, Avgelis A, Katis NI, Maliogka VI (2017) Insights into the etiology of polerovirus-induced pepper yellows disease. Phytopathology 107:1567–1576
MacKenzie DJ, McLean MA, Mukerji S, Green M (1997) Improved RNA extraction from woody plants for the detection of viral pathogens by reverse transcription-polymerase chain reaction. Plant Dis 81:222–226
Maina S, Edwards OR, Jones RAC (2016) First complete genome sequence of Pepper vein yellows virus from Australia. Genome Announc 4:e00450-e1416
Makkouk KM, Comeau A (1994) Evaluation of various methods for the detection of barley yellow dwarf virus by the tissue-blot immunoassay and its use for virus detection in cereals inoculated at different growth stages. Eur J Plant Pathol 100:71
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003–vev003
Mayo MA, D’Arcy CJ (1999) Family Luteoviridae: a reclassification of Luteoviruses. CABI publishing, Wallingford
Mayo MA, Miller WA (1999) The structure and expression of luteovirus genomes. In: Smith HG, Barker H (eds) The Luteoviridae. CAB International, Wallingford, pp 23–42
Mayo MA (2002) virology division news: ICTV at the Paris ICV: results of the plenary session and the binomial ballot. Arch Virol 147:2254–2260
Minh BQ, Hahn MW, Lanfear R (2020) New methods to calculate concordance factors for phylogenomic datasets. Mol Biol Evol 37:2727–2733
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9:e108277–e108277
New S-A, van Heerden SW, Pietersen G, Esterhuizen LL (2016) First report of a turnip yellows virus in association with the brassica stunting disorder in South Africa. Plant Dis 100:2341
Newbert M (2016) The genetic diversity of Turnip yellows virus in oilseed rape (Brassica napus) in Europe, pathogenic determinants, new sources of resistance and host range. Doctoral dissertation, University of Warwick, Coventry. http://webcat.warwick.ac.uk/record=b2869367~S1
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2014) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Pagán I, Holmes EC (2010) Long-term evolution of the luteoviridae: time scale and mode of virus speciation. J Virol 84:6177–6187
Passmore BK, Sanger M, Chin LS, Falk BW, Bruening G (1993) Beet western yellows virus-associated RNA: an independently replicating RNA that stimulates virus accumulation. P Natl Acad Sci 90:10168–10172
Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Xe R, Hemmer O, Kretsch T, Richard KE, Genschik P, Ziegler-Graff V, Xe R (2006) F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci USA 103:1994–1999
Peter KA, Liang D, Palukaitis P, Gray SM (2008) Small deletions in the potato leafroll virus readthrough protein affect particle morphology, aphid transmission, virus movement and accumulation. J Gen Virol 89:2037–2045
Peter KA, Gildow F, Palukaitis P, Gray SM (2009) The C terminus of the polerovirus P5 readthrough domain limits virus infection to the phloem. J Virol 83:5419–5429
Prüfer D, Kawchuk LM, Rohde W (2006) Polerovirus ORF0 genes induce a host-specific response resembling viral infection. Can J Plant Path 28:302–309
Sánchez PAG, Mesa HJ, Montoya MM (2016) Next generation sequence analysis of the forage peanut (Arachis pintoi) virome. Revista Facultad Nacional de Agronomía Medellín 69:7881–7891
Sanger M, Passmore B, Falk BW, Bruening G, Ding B, Lucas WJ (1994) Symptom severity of beet western yellows virus strain ST9 is conferred by the ST9-associated RNA and is not associated with virus release from the phloem. Virology 200:48–55
Schliephake E, Graichen K, Rabenstein F (2000) Investigations on the vector transmission of the Beet mild yellowing virus (BMYV) and the Turnip yellows virus (TuYV). Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz 107:81–87
Schmitz J, Stussi-Garaud C, Tacke E, Prüfer D, Rohde W, Rohfritsch O (1997) In situlocalization of the putative movement protein (pr17) from Potato leafroll luteovirus (PLRV) in infected and transgenic potato plants. Virology 235:311–322
SeedWorld (2019) Turnip yellows virus: the underestimated threat in oilseed rape cultivation. https://seedworld.com/turnip-yellows-virus-the-underestimated-threat-in-oilseed-rape-cultivation/
Shahraeen N, Farzadfar S, Lesemann D-E (2003) Incidence of viruses infecting winter oilseed rape (Brassica napus ssp. oleifera) in Iran. J Phytopathol 151:614–616
Sharman M, Lapbanjob S, Sebunruang P, Belot JL, Galbieri R, Giband M, Suassuna N (2015) First report of Cotton leafroll dwarf virus in Thailand using a species-specific PCR validated with isolates from Brazil. Australas Plant Dis Notes 10:1–4
Shukla DD, Ward CW (1988) Amino acid sequence homology of coat proteins as a basis for identification and classification of the potyvirus group. J Gen Virol 69:2703–2710
Simon-Loriere E, Holmes EC (2011) Why do RNA viruses recombine? Nat Rev Microbiol 9:617–626
Smirnova E, Firth AE, Miller WA, Scheidecker D, Brault V, Reinbold C, Rakotondrafara AM, Chung BYW, Ziegler-Graff V (2015) Discovery of a small non-AUG-initiated ORF in poleroviruses and luteoviruses that is required for long-distance movement. PLoS Pathog 11:e1004868
Stevens M, Smith HG, Hallsworth PB (1995) Detection of the luteoviruses, beet mild yellowing virus and beet western yellows virus, in aphids caught in sugar-beet and oilseed rape crops, 1990–1993. Ann Appl Biol 127:309–320
Stevens M, McGrann G, Clark B, Authority H (2008) Turnip yellows virus (syn Beet western yellows virus): An emerging threat to European oilseed rape production. Research Review 69 HGCA. Available via: http://www.hgca.com/document.aspx?fn=load&media_id=4473&publicationId=4579
van den Heuvel JF, Bruyère A, Hogenhout SA, Ziegler-Graff V, Brault V, Verbeek M, van der Wilk F, Richards K (1997) The N-terminal region of the luteovirus readthrough domain determines virus binding to Buchnera GroEL and is essential for virus persistence in the aphid. J Virol 71:7258–7265
van der Wilk F, Verbeek M, Dullemans AM, van den Heuvel JFJM (1997) The genome-linked protein of potato leafroll virus is located downstream of the putative protease domain of the ORF1 product. Virology 234:300–303
van Leur JAG, Aftab M, Manning W, Bowring A, Riley MJ (2013) A severe outbreak of chickpea viruses in northern New South Wales, Australia, during 2012. Australas Plant Dis Notes 8:49–53
Wang F, Wu QF, Zhou BG, Gao ZL, Xu DF (2015) First Report of Turnip yellows virus in Tobacco in China. Plant Dis 99:1870–1870
Worobey M, Holmes EC (1999) Evolutionary aspects of recombination in RNA viruses. J Gen Virol 80:2535–2543
Xiang H-Y, Dong S-W, Shang Q-X, Zhou C-J, Li D-W, Yu J-L, Han C-G (2011) Molecular characterization of two genotypes of a new polerovirus infecting brassicas in China. Arch Virol 156:2251–2255
Yoshida N, Tamada T (2019) Host range and molecular analysis of Beet leaf yellowing virus, Beet western yellows virus-JP and Brassica yellows virus in Japan. Plant Pathol 68:1045–1058
Zhang X-Y, Xiang H-Y, Zhou C-J, Li D-W, Yu J-L, Han C-G (2014) Complete genome sequence analysis identifies a new genotype of brassica yellows virus that infects cabbage and radish in China. Arch Virol 159:2177–2180
Ziegler-Graff V, Brault V, Mutterer JD, Simonis MT, Herrbach E, Guilley H, Richards KE, Jonard G (1996) The coat protein of beet western yellows luteovirus is essential for systemic infection but the viral gene products P29 and P19 are dispensable for systemic infection and aphid transmission. Mol Plant Microbe In 9(6):501–510
Acknowledgements
This study was funded by the Queensland Department of Agriculture and Fisheries and the Australian Grains Research and Development Corporation projects DAN00202, DAQ00186 and DAQ00154. We thank Dr. Joop Van Leur and Dr. Brenda Coutts for discussions about the project and assistance with sample collection, and Dr. Paul Campbell for discussions about the results.
GenBank accession numbers MT586571– MT586598 and MT642436–MT642445.
Funding
This study was funded by the Queensland Department of Agriculture and Fisheries and the Australian Grains Research and Development Corporation (projects DAN00202, DAQ00186 and DAQ00154).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare there are no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Jesús Navas-Castillo.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Filardo, F., Nancarrow, N., Kehoe, M. et al. Genetic diversity and recombination between turnip yellows virus strains in Australia. Arch Virol 166, 813–829 (2021). https://doi.org/10.1007/s00705-020-04931-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04931-w